This blog post targets researchers concerned about pollution and the environment, focusing on using alternative eco-friendly methods to remediate these contaminated environments. It also targets microbiologists and biotechnologists, encouraging them to leverage microorganisms for environmental benefits.
Hello, I am Asia from Sudan. I am very grateful for the opportunity to pursue my master’s degree at the University of Bergen.
This semester, I have learned many new skills that I am eager to apply to my thesis research. The BIO299 course has provided an excellent hands-on experience, allowing me to practice these skills and prepare for my thesis work. It has been particularly valuable for lab time management, handling errors, and adhering to the research plan as closely as possible.
In addition to practicing lab work and gaining new writing and communication skills, the aim of this project is to extract and characterize a potential cyanide hydrolase enzyme from Bacillus haynesii NRRL B-41327. This strain, which is well-suited for bioremediation of cyanide-contaminated environments, was isolated from a mining site in northeastern Sudan.
Cyanide can be found in different forms in the environment, either naturally or due to industrial activities such as silver and gold mining. Cyanide contamination is increasing in the environment, posing a risk of introduction into the human body.
Both chemical and bioremediation methods can be used to remove cyanide from the environment. Bioremediation is considered cheaper and more effective, especially when it comes to degradation byproducts, compared to chemical methods.
In our project, we successfully expressed and purified a putative cyanide hydrolase enzyme from Bacillus haynesii NRRL B-41327, using Escherichia coli BL21-Gold (DE3) as an expression host for our codon-optimized gene in the pET-28b expression plasmid.
Our results are consistent with the findings of Chin et al. (2007) and Tsai et al. (2006), indicating that our protein belongs to the CN-hydrolase superfamily as shown in Figure 1A.
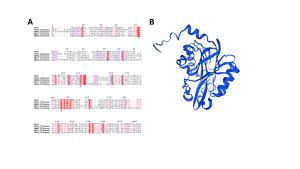
Figure 1A: The protein sequences from our Bacillus haynesii NRRL B-41327 strain were aligned with CN-hydrolysing enzymes from various species including Saccharomyces cerevisiae, Caenorhabditis elegans, Xanthomonas campestris, and Agrobacterium sp. These sequences, obtained from the PDB, were aligned using the MAFFT tool and visualized with ESPript 3.0. Conserved catalytic amino acids in the active sites, indicated by a blue star at positions 42, 111, and 145, were identified based on the studies by (Chin et al., 2007) and (Tsai et al., 2006). Figure 1B: 3D structure of the target protein as modeled by the Swiss-Model.
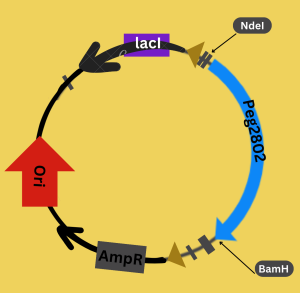
Figure 2: We constructed our plasmid using GeneScript, with codon optimization for E. coli. The constructed plasmid contains our target protein Peg2802, the ampicillin resistance gene for selection (AmpR), the lacI gene (Lac Repressor Gene) to induce protein expression with IPTG, the origin of replication (ori), and the restriction sites NdeI and BamHI.

Figure 3: This figure shows the successful protein expression in cell transformed the expression plasmid (R1, R2) compared to a control sample (Control) that lacks the target protein. The target protein appears as a band of approximately 30 kDa in the SDS-PAGE analysis gel.

Figure 4: The protein was purified using Immobilized Metal Affinity Chromatography (IMAC) performed on an ÄKTA Start with a 5 mL HisTrap FF column. «PS» shows the protein standard, «Total» shows the total cell sample, «Soluble» is the sonicated sample after centrifugation, and «Flow Through (Fth)» shows the washed unbound target protein. Fractions from F14 to F28 contain our target protein. The target protein appears as a band of approximately 30 kDa in the SDS-PAGE analysis gel.
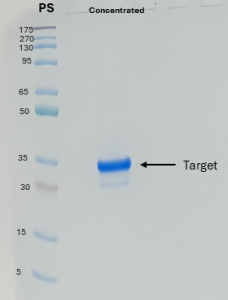
Figure 5: The protein was concentrated using Amicon® Ultra-15 10K Centrifugal Devices while exchanging the buffer. The eluted fractions from the IMAC step (fractions F14 to F28) were combined and then concentrated. The target protein appears as a band of approximately 30 kDa in the SDS-PAGE analysis gel.
We have successfully expressed the potential cyanide hydrolase enzyme in soluble form and purified it to near homogeneity. Nevertheless, its activity still needs to be confirmed through biochemical tests.
References:
Jaszczak, E., Polkowska, Ż., Narkowicz, S., Namieśnik, J., 2017. Cyanides in the environment—analysis—problems and challenges. Environ Sci Pollut Res 24, 15929–15948. https://doi.org/10.1007/s11356-017-9081-7
Tiong, B., mohd bahari, Z., Lee, N., Jaafar, J., Ibrahim, Z., Shahir, S., 2014. Cyanide Degradation by Pseudomonas pseudoalcaligenes Strain W2 Isolated from Mining Effluent. Sains Malaysiana. https://doi.org/10.17576/jsm-2015-4402-10
Kandasamy, S., Dananjeyan, B., Krishnamurthy, K., Benckiser, G., 2015. Aerobic cyanide degradation by bacterial isolates from cassava factory wastewater. Braz J Microbiol 46, 659–666. https://doi.org/10.1590/S1517-838246320130516
Chin, K.-H., Tsai, Y.-D., Chan, N.-L., Huang, K.-F., Wang, A.H.-J., Chou, S.-H., 2007. The crystal structure of XC1258 from Xanthomonas campestris: a putative procaryotic Nit protein with an arsenic adduct in the active site. Proteins 69, 665–671. https://doi.org/10.1002/prot.21501
Tsai, Y.-D., Chin, K.-H., Shr, H.-L., Gao, F.P., Lyu, P.-C., Wang, A.H.-J., Chou, S.-H., 2006. Cloning, crystallization and preliminary X-ray study of XC1258, a CN-hydrolase superfamily protein from Xanthomonas campestris. Acta Crystallogr Sect F Struct Biol Cryst Commun 62, 999–1002. https://doi.org/10.1107/S1744309106035433