Mackerel, one of the most important pelagic fish inhabiting in both temperate and tropical seas, mostly living along the coast or offshore in the oceanic environment and may predate on larval and juvenile herring when they are adult. So we want to map and quantify grazing pressure on spring-spawning herring (Clupea harengus) larvael.
However, I want to talk more about the molecular techniques to detect the herring predation rate by Mackerel in different areas of atlantic ocean. The whole process starts with first identifying the sampling sites where different phenomena take place in terms of herring larvae consumed by the pelagic fish Mackerel.
Our study choose different fjords and ocean flows at different places around the geographic region of Norway. Mackerel from different sampling sites were recruited for the study to see how they consumed herring larvae.
Coming to the molecular lab, Mackerel stomach was harvested and whole genomic DNA were extracted following DNA extraction protocol. You must be careful in this step to avoid any potential contamination with possibly yours, mine or any other DNA from unexpected sources.
As we earlier designed our primers specific to the conserved region of 16s ribosomal RNA of atlantic herring (Clupea herengus), we can use this primer to enhance by PCR the tiny amount of DNA into sample. The primer design takes place using bioinformatics tools and online open database search.
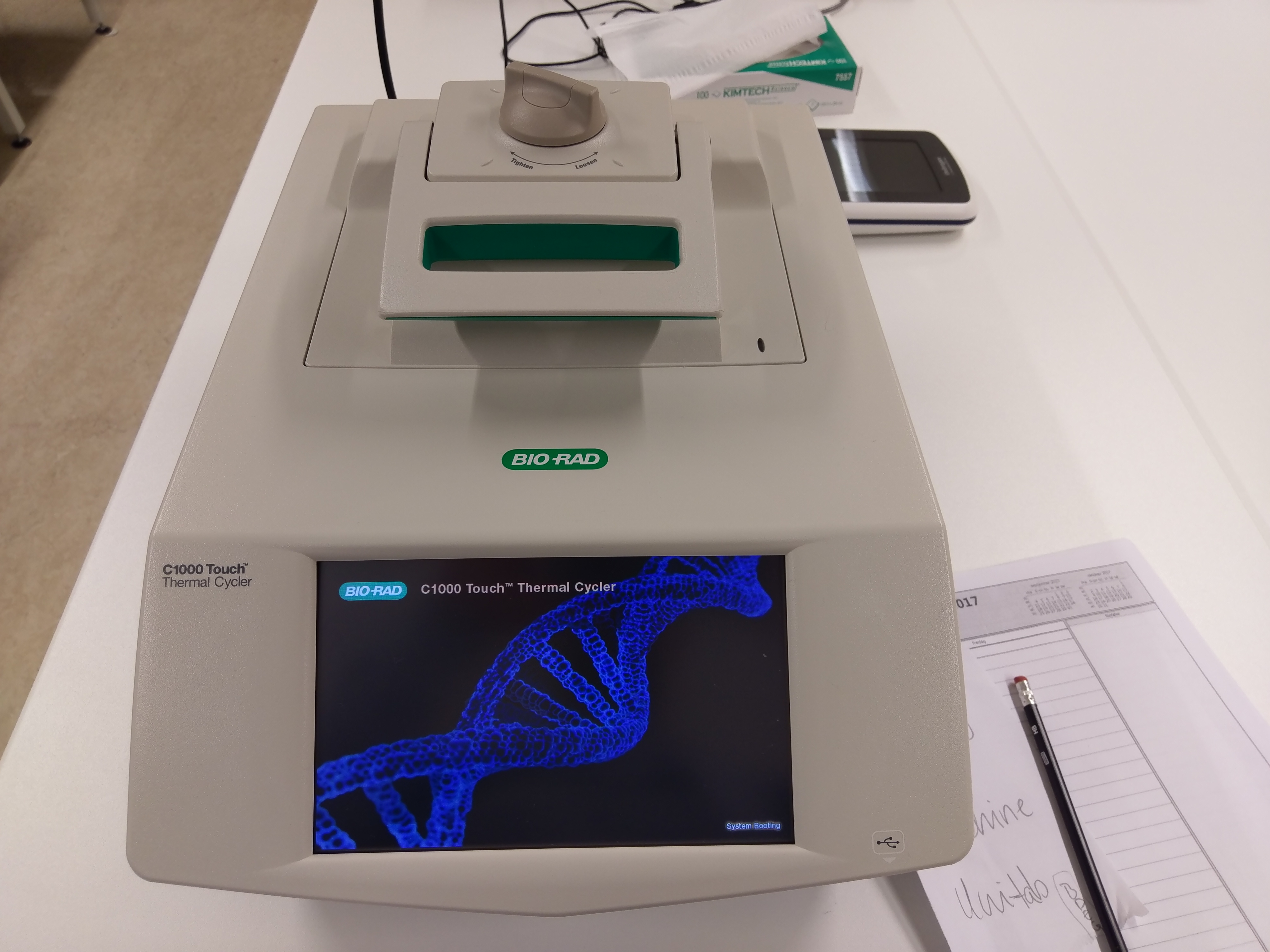
Nevertheless, we need to run a specificity test of primer before we decide on optimized condition for our PCR. We used here digital droplet PCR (ddPCR) that measures the fraction of negative replicates to determine absolute copies.
Figure 1: Digital Droplet PCR setup
In optimization, we consider our variables- primer concentration, primer annealing temperature in combination with different dilution of template DNA including positive and negative controls. Different dilution can be used to test sensitivity of PCR.
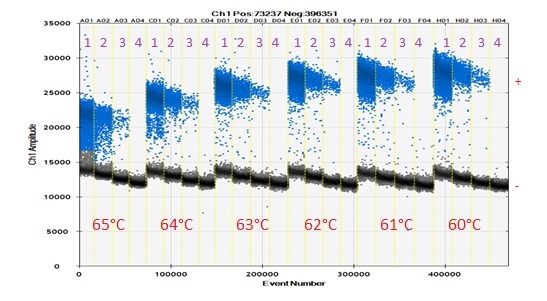
Figure 2: The gradient PCR reaction at different annealing temperature for serial dilution of templates to 102 fold, 103 fold and 104 fold (1-3) with a blank (4). Blue indicates positive droplets and the black negative. At 65°C , the resolution between positive and negative droplets were worst and at 60°C, it is the best with minimum rain of droplets. Hence, 60 °C can be considered perfect annealing temperature for this PCR program.
By specificity test, we confirm whether our selected primer at defined concentration and annealing temperature doesn’t bind with unintended DNA template (nonspecific binding). PCR reaction with different target templates from closely related organisms is set for determining primers specificity.
After optimization, sensitivity and specificity test, we set our PCR with all DNA templates from Mackerel collected from various sources. Some were showing high DNA copies of herring and some showed negative. This picture gives us an idea for mapping and quantification of grazing pressure on spring-spawning herring (Clupea harengus) larvael.