Hi, my name is Anton Melbye and I’m doing research practice this fall. My project was about identifying proteins associated with old nuclear pores in budding yeast. In simpler words: I worked with transformed yeast strains and took pictures of the nuclear pore complexes. I transformed the strains with homologous recombination to add fluorescence in different proteins. My work mostly consisted of growing yeast strains, making media, and doing fluorescence microscopy.
I spent many hours in the lab, and I guess you could say that yeast has been my best friend the last couple of months. Yeast requires some supervision since I always had to check if they had grown enough, if they are overgrown or if I even grew the correct strains. They don’t give much in return, but it was still fun to do take care of them.
My project was important because there is still a lot to learn about the proteins composing the nuclear pore complexes (NPC) and their function. My supervisor Evgeny Onishchenko has been working with nuclear pore complexes for quite some time and helped me along the way. I chose to only work on proteins associated with old pores in yeast because it would be too time-consuming to take on both old and new proteins. The proteins I settled with were ESC1, PML39, PRE4, SAC3, STS1 and ULP1. They were chosen by recommendation from my supervisor as they are promising candidates.
Acquired skills during this project:
- PCR
- Gel electrophoresis
- Primer design
- Making yeast media and growing yeast cultures
- Fluorescence microscopy
- Quantitative image analysis
- Genetic transformation
What are nuclear pore complexes?
Nuclear pore complexes (NPCs) are transport channels in the nuclear membrane. They allow macromolecules to move between the nucleus and the cytoplasm. NPCs are composed of proteins called nucleoporins. Some structures of the NPC can survive for a long time and can accumulate damage. Many neurodegenerative disorders are associated with NPC dysfunction, which affirms the importance of this project: to get more knowledge about NPCs.
Preparation and workflow:
Mainly, the project workflow has been about growing various strains of yeast and modifying them genetically with fluorescence for microscopy.
Growing yeast on selection media:
Ordering primers:
It was fun to get hands on experience with genetics and primer design. I have previously learned about this in class, but only theoretically.
Using a plasmid editor program called APE, I modified the open reading frames of the selected proteins to add mCherry for red fluorescence. I picked a forward and a reverse primer from the modified loci and ordered them. The primers were used to make a “cassette” which is used to transform cells by homologous recombination.
PCR:
I used PCR to accumulate the cassettes used in transformation. The PCR product then had to be tested using gel electrophoresis. Luckily, I had done both PCR and gel electrophoresis before and this went smoothly.

Gel electrophoresis results for the PCR products for each strain. One PCR product was not successful but was redone with higher quantities of DNA successfully.
Transforming yeast strains:
The PCR products (Cassettes) were used to transform the strains by homologous recombination. For this, I used a transformation mix, which is standardized and easy to make. To my surprise, the transformation mix I made had salmon sperm DNA in the recipe, which I think was a bit funny. I never fully understood the importance of salmon sperm in my yeast transformation mix, but it worked so I can’t complain.
Then, the yeast strains were grown on selection media to only grow the transformed strains. Each strain had to be tested in fluorescence microscopy to check if the transformation was successful. The yeast
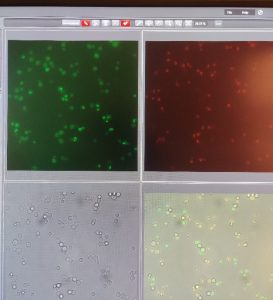
strains used to test old nuclear pores has no green fluorescence before recombination, but will have red fluorescence because of the inserted mCherry cassette.The red fluorescence shows that this was a successful transformation. Some green fluorescence is present because of stochastic recombination, allowing GFP to be expressed.
Microscopy and image processing:
I had to take a lot of images of each yeast strain to get enough data. In total I think I captured around 400 images using the fluorescence microscope. The images were run through a computer program which measured light intensity in the nuclear envelopes. We then used this data to see if the selected proteins preferred old pores.

This is a representative image I made to show how we can stack images with different colored proteins on top of each other to see if there is a correlation between the two. If the proteins localize together, the cells will have fluorescence in the same pores.
Results
My findings in this project were that Esc1, Pml39, Pre4 and Ulp1 are good candidates for proteins that prefer old pores. The data I gathered is not conclusive, but it gives good evidence for future research. Esc1, Pml39, Pre4 and Ulp1 could potentially be used as biomarkers for old NPCs. My supervisor and his team will continue gathering evidence for proteins like these and will do more extensive projects to map the maturation kinetics of NPCs.
Final thoughts:
When I first started this project, I thought it was a lot more than I could take on and felt overwhelmed with work. However, after taking some time to figure out how it all worked, it was achievable. With good help and supervision from my supervisor, most of the work went smoothly.
My work also makes a good starting point for other students participating on this project. Next person can use a lot of the information we gathered and improve many of the methods to get more information about nucleoporins and NPCs.
The most exciting aspect of this project is contributing to actual research. This is valuable for my education and future career. I am grateful to be part of such a big project and it made me a lot more interested in this line of work. I greatly recommend participating in bio299 to get unique experiences during your biology degree.