Deep beneath the ocean’s surface, hidden ecosystems thrive in the darkness. Hydrothermal vents release water rich in chemicals that allow life to adapt to extreme conditions. In these unique environments, the primary producers don’t use sunlight, instead microbes use the strong chemical gradient to support a complex ecosystem. Some of the microbes inhabiting the deep sea consume methane, a potent greenhouse gas as an energy source. They are single-cell archaea that team up with bacteria in a relationship known as a synthropic consortium. This remarkable partnership regulates methane release into the environment.
My investigation focused on a sample collected from Loki’s Castle, a methane-rich hydrothermal vent field in the Arctic Mid-Ocean Ridge (AMOR). This sample, preserved in a bottle for nearly six years, mimicking the original environment – cold, dark, oxygen-free, and initially filled with methane gas. But how do we reveal the microbial inhabitants hidden in this bottle? This is where metagenomics comes in, a powerful tool that lets us peer into microbial communities without needing to isolate each individual organism. From a small portion of the sample, we could analyze all the DNA at once, revealing the incredible diversity of microbes present. However, it’s a computationally demanding process, requiring access to powerful supercomputers operated remotely.
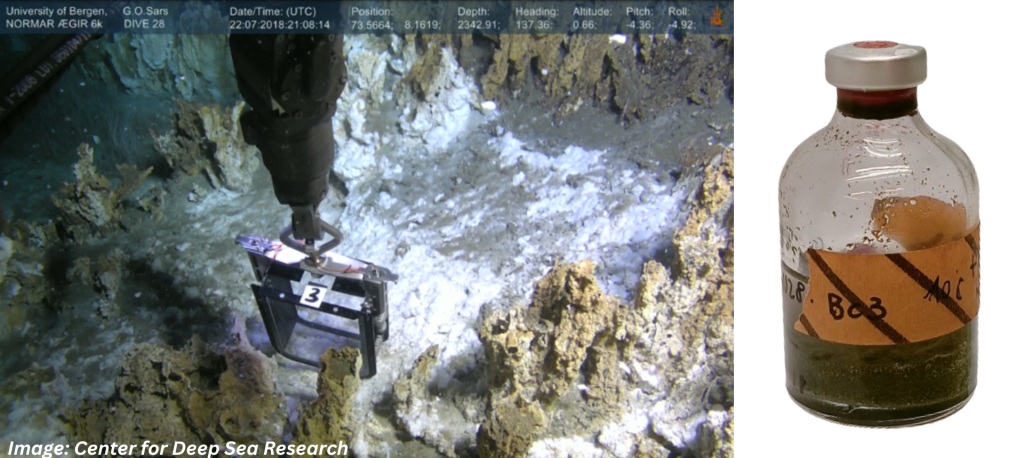
Images: The sample location in the sediments Loki’s Castle vent field in the Arctic Mid-Ocean Ridge (AMOR), and the serum bottle incubated a low temperature for almost six years.
Despite the years spent in a bottle, the sample surprised us with over 200 types of different microbes! Most of them new species and the fascinating partnership that consumes methane as a dominant metabolism. The key archaea are known as ANaerobic MEthane oxidizing Archaea or ANME. They usually partner with the Sulfate Reducing Bacteria or SRB. Here’s how it works: ANME consumes methane, but in the process generates electrons. SRBs are happy to accept these electrons and use them to transform sulfate into sulfide, their own energy source. Interestingly, we discovered high concentrations of sulfide indicating that these partnerships between ANME and SRB were indeed active within the bottle. However, sulfide is toxic at such levels, so it was a surprise to see the ANME cells appeared to be thriving under the microscope! In addition, partnerships between ANME and SRB were generating organic matter (primary producers) that supported the growth of other organisms. Increasing the metabolic diversity of the sample.
The most amazing aspect of our findings was the immense number of novel species! One particular SRB had never been documented at such low temperature, challenging our understanding of these microbial communities. These findings raise a multitude of questions: How much remains still unknown about these deep-sea ecosystems? What potential benefits could these microbes offer in mitigating global methane emissions? Conversely, what threats might they face due to climate change or human activities?
This research highlights the limitations of our knowledge about deep-sea marine ecosystems. Future research efforts, such as analyzing the genomes of these microbes in more detail, may help us to discover new enzymes or metabolic pathways with potential applications in biotechnology or bioremediation, including mediating with methane emissions. By unraveling the complex interactions within these microbial communities, we gain a deeper understanding of the diversity and resilience of life on Earth as a system.
This research learning journey has been amazing. I would like to express my sincere gratitude to my supervisor and colleagues for their support throughout this project.